IMIB publications
filler
For a complete publication list please use one of the links above
*corresponding author, ± co-first author
PhotoFiTT: A Quantitative Framework for Assessing Phototoxicity in Live-Cell Microscopy Experiments, ±Del Rosario M, ±Gómez-de-Mariscal E, Morgado L, Portela R, Jacquemet G, Pereira PM*, Henriques R*, Nature Communications (2025).
What is this about? PhotoFiTT is a novel quantitative framework that assesses phototoxicity in live-cell microscopy experiments by combining a standardized protocol with advanced image analysis. It leverages machine learning and cell cycle dynamics to analyze mitotic timing, cell size changes, and cellular activity in response to controlled light exposure, enabling researchers to establish quantitative benchmarks for acceptable levels of photodamage and optimize imaging protocols
MicroMundo@Oeiras: citizen science promoting antibiotic stewardship , discovery of new antimicrobials, and monitoring of soil resistance, ±Henriques B, ±Brito B, ±Barreiros C, ±Galvão G, ±Coito J, ±Lourinho S, ±Mil-Homens T, ±Maciel V, ±Queijinho A, ±Cardoso B, ±Maria L, ±Vatova M, ±Bastos R, ±Prata SC, Martins LO, Ramalho R, Silva A, Brigadeiro E, Leão MJ*, Pereira PM*, Durão P*, FEMS Microbiology Letters (2025).

What is this about? The Micromundo@Oeiras project is implemented by ITQB NOVA within the scope of the Active Citizenship Program – Ciência + Cidadã (C+C), in close collaboration with the Municipality of Oeiras. Micromundo@Oeiras is the result of a collaboration with MicroMundo@UPorto, implemented by Universidade do Porto as part of the international project Small World Initiative (SWI)/Tiny Earth). We explore the biodiversity of local soil, searching for new microbes that can produce antibiotics. This hands-on approach is a powerful way to educate young people about the AMR crisis. We expanded our research beyond just finding antibiotic producers. We have developed methods to also measure the frequency of antibiotic-resistant bacteria in the very same soil samples. Specifically, we test for resistance to common antibiotics like amoxicillin, tetracycline, and ciprofloxacin. Our initial findings are fascinating: it appears that bacteria resistant to these antibiotics are more common in our local soil than the bacteria that can produce new ones. This surprising result has been a stimulating discovery for our citizen scientists and has opened up new avenues for our research. We have created a public database. We will continuously update it with our findings on AMR frequency in soil bacteria, creating a novel and valuable source of data from unconventional places like our local parks and gardens.
Efficiently accelerated bioimage analysis with NanoPyx, a Liquid Engine-powered Python framework Saraiva BM, Cunha IM, Brito AD, Follain G, Portela R, Haase R, Pereira PM, Jacquemet G, Henriques R, Nature Methods (2025).
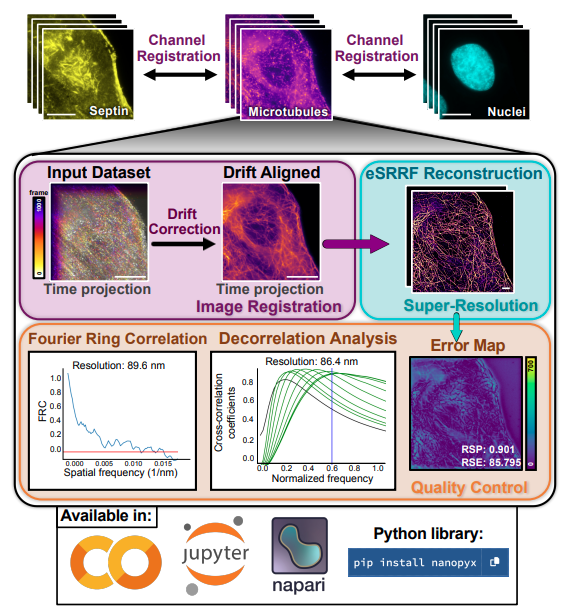
What is this about? NanoPyx is a cutting-edge bioimage analysis framework that accelerates the processing of complex microscopy datasets. Its adaptive machine learning system, the Liquid Engine, optimizes performance by dynamically generating and benchmarking multiple code variations for each task, achieving over 10-fold speed improvements. NanoPyx offers accessibility through a Python library, Jupyter notebooks, and a napari plugin, catering to users of all coding proficiency levels.
Como dialogar com quem não quer ouvir: para lá da polarização e da desinformação, Marcelo G*, Pereira PM*, Sanchez A*, Freitas MS*, Academia de Ciências de Lisboa (2023).
What is this about? We explore strategies for effective communication in an era marked by polarization and the spread of misinformation. We aim to propose techniques to foster meaningful dialogue, even with individuals who may be resistant to opposing viewpoints or facts. By moving beyond the barriers created by entrenched beliefs and the influence of false information, the book aims to provide tools for building bridges and promoting understanding in challenging conversational landscapes.
DeepBacs for multi-task bacterial image analysis using open-source deep learning approaches, Spahn C, Laine RF, Gómez-de-Mariscal E, Pereira PM , von Chamier L, Conduit M, Pinho MG, Jacquemet G, Holden S, Heilemann M, Henriques R, Communications Biology (2022).

What is this about? We suggest how to use a range of state-of-the-art artificial neural networks to analyze bacterial microscopy images using the recently developed ZeroCostDL4Mic platform. We generated a database of image datasets used to train networks for various tasks, including segmenting bright field and fluorescence images of different bacterial species, classifying growth stages in time-lapse data, and profiling antibiotic-treated cells. We also showcase how deep learning can enhance low-phototoxicity live-cell microscopy through denoising, artificial labelling of cell membranes, and predicting super-resolution images. The purposefully-built database aids in training novice users, enabling them to quickly explore data analysis through deep learning, fostering the efficient application of these techniques in microbiology and the creation of tools for bacterial cell biology and antibiotic research.
Single-molecule super-resolution imaging of T-cell plasma membrane CD4 redistribution upon HIV-1 binding, Yuan Y, Jacobs C, Garcia IL, Pereira PM , Lawrence SP, Laine RF, Marsh M, Henriques R, Viruses (2021).
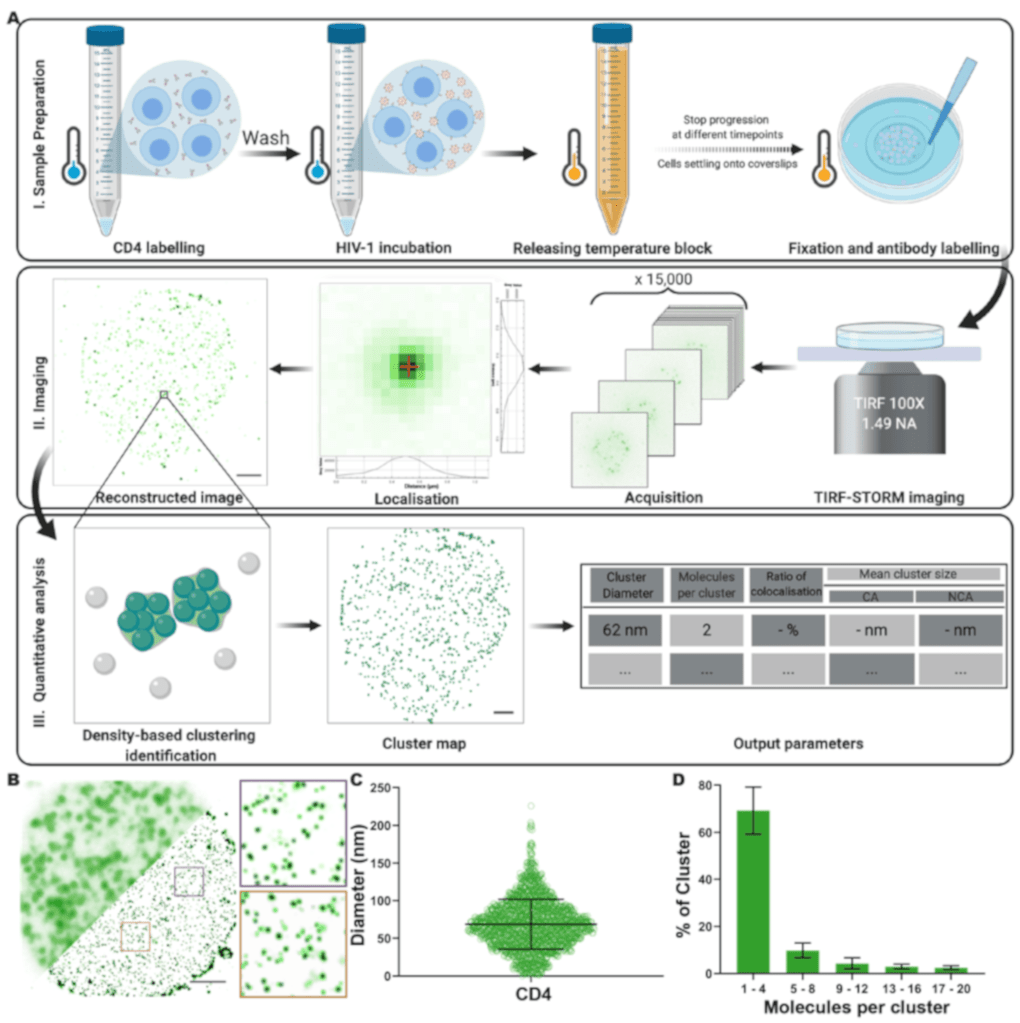
What is this about? We use single-molecule super-resolution imaging to quantitatively examine the nanoscale reorganization of CD4 receptors on CD4+ T cell surfaces upon HIV-1 binding. We show that CD4 molecules are primarily distributed as individual molecules or small clusters in the absence of virus. Following HIV-1 or recombinant gp120 binding, we observe a local increase in cluster diameter and molecule number for virus-associated CD4 clusters. This study provides a robust methodology for characterizing plasma membrane receptor organization and quantifies CD4 redistribution triggered by HIV-1 on host cells.
Super-Beacons: Open-Source Probes With Spontaneous Tuneable Blinking Compatible With Live-Cell Super-Resolution Microscopy, ±Pereira PM, ±Gustafsson N, Marsh M, Mhlanga MM, Henriques R, Traffic (2020).
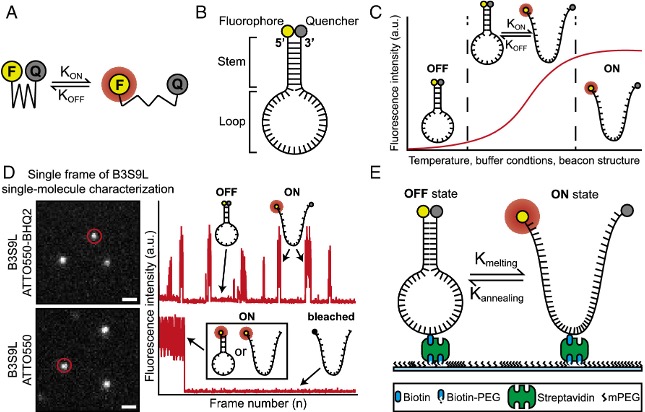
What is this about? We developed a new class of DNA-based open-source super-resolution probes named Super-Beacons, which exhibit spontaneous photoswitching based on a self-quenching mechanism. Super-Beacons enable live-cell compatible super-resolution microscopy without the need for high-intensity illumination or toxic imaging buffers. We demonstrated the potential of these probes by imaging interferon-inducible transmembrane proteins (IFITMs) at sub-100 nm resolutions. The photoswitching kinetics of Super-Beacons can be tuned structurally, thermally, and chemically, making them a versatile tool for live-cell super-resolution imaging.
Nuclear pores as versatile reference standards for quantitative superresolution microscopy, Thevathasan JV, Kahnwald M, Cieslinski k, Hoess P, Peneti SK, Reitberger M, Heid D, Kasuba KC, Hoerner SJ, Li Y, Wu YL, Mund M, Matti U, Pereira PM , Henriques R, Nijmeijer-Winter B, Kueblbeck M, Sabinina VJ,Ellenberg J, Ries J, Nat. Methods (2019).

What is this about? We exploited the stereotypic arrangement of proteins in the nuclear pore complex as in situ reference structures to characterize the performance of various superresolution microscopy modalities. By endogenously labeling the nucleoporin Nup96 with mEGFP, SNAP-tag, HaloTag, or the photoconvertible fluorescent protein mMaple in four genome-edited cell lines, we demonstrated their use as three-dimensional resolution standards for calibration and quality control, to quantify absolute labeling efficiencies, and as precise reference standards for molecular counting. Our cell lines will enable the broader community to assess the quality of their microscopes and labels, and to perform quantitative, absolute measurements.
Fix your membrane receptor imaging: Actin cytoskeleton and CD4 membrane organization disruption by chemical fixation, ±Pereira PM*, ±Albrecht D*, Culley S, Jacobs C, Marsh M, Mercer J, Ricardo Henriques*, Frontiers in Immunology (2019).

What is this about? We explored how different fixation approaches that disrupt or preserve the actin cytoskeleton affect membrane protein organization, using CD4 as a model. We showed that fixation-mediated disruption of the actin cytoskeleton correlates with changes in CD4 membrane organization. We highlight how these artifacts are easy to overlook and how careful sample preparation is essential for extracting meaningful results from super-resolution microscopy. Although the actin cytoskeleton and protein-protein interactions are important for membrane protein organization, many SMLM studies focus on imaging unknown structures and distributions of proteins without a known organization. When performing essential protocol optimization, preservation of the overall cellular structure should be a priority.
Automating multimodal microscopy with NanoJ-Fluidics, ±Almada P, ±Pereira PM, Culley S, Caillol G, Boroni-Rueda F, Dix CL, Charras G, Baum B, Laine RF, Leterrier C, Henriques R, Nat. Communications (2019).

What is this about? We developed NanoJ-Fluidics, an open-source framework that enables precise automated fluid exchange in commonly used sample chambers, making complex multimodal imaging protocols easy to implement on any microscope. NanoJ-Fluidics is based on low-cost Lego hardware controlled by ImageJ-based software. We demonstrate its applicability in multiple experimental contexts, including event-driven fixation, in-situ correlative live-to-fixed super-resolution imaging, and multimodal STORM/DNA-PAINT experiments. NanoJ-Fluidics makes complex multimodal microscopy experiments accessible to researchers, regardless of their background, and opens the door to the development of novel assays and imaging approaches.
NanoJ: a high-performance open-source super-resolution microscopy toolbox, Laine RF, Tosheva KL, Gustafsson N, Gray RDM, Almada P, Albrecht D, Risa GT, Hurtig F, Lind ̊as AC, Baum B, Mercer J, Leterrier C, Pereira PM* , Culley S*, Ricardo Henriques*, J. Phys. D (2019).
What is this about? We developed NanoJ, a high-performance open-source toolbox for super-resolution microscopy designed to combine powerful image analysis capabilities with ease of use. NanoJ includes tools for spatio-temporal alignment of raw data (NanoJ-Core), super-resolution image reconstruction (NanoJ-SRRF), image quality assessment (NanoJ-SQUIRREL), structural modeling (NanoJ-VirusMapper), and control of the sample environment (NanoJ-Fluidics). We continue to expand NanoJ with new tools to improve quantitative data analysis and measure the reliability of fluorescent microscopy experiments. The software modules of NanoJ are part of our ImageJ/Fiji image analysis framework, making it accessible to a wide range of researchers.
Septins Recognise Bacterial Cell Division for Host Defence, Krokowski S, Lobato-Márquez D, Chastanet A, Pereira PM , Angelis D, Galea1 D, Larrouy-Maumus G, Henriques R, Spiliotis ET, Carballido-López R, Mostowy S, Cell Host & Microbe (2018).
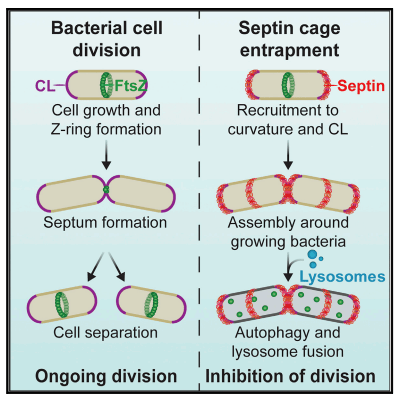
What is this about? We discovered that septins, a component of the host cytoskeleton, recognize the cell division site of intracellular bacterial pathogens such as Shigella. We found that septin recruitment to dividing bacterial cells is promoted by the presence of cardiolipin, a curvature-specific phospholipid enriched at the bacterial division site. Once recruited, septins assemble into cage-like structures around growing bacterial cells, restricting their proliferation by targeting them to autophagy and lysosomal fusion. Our findings reveal a novel mechanism by which the host cell can sense and defend against intracellular bacterial infection by recognizing the unique geometry and membrane composition associated with bacterial cell division.
Quantitative mapping and minimization of super-resolution optical imaging artifacts, Culley S, Albrecht D, Jacobs C, Pereira PM, Leterrier C, Mercer J, Ricardo Henriques, Nat. Methods (2018).

What is this about? We developed NanoJ-SQUIRREL, an ImageJ-based analytical approach that provides quantitative assessment of super-resolution image quality. SQUIRREL compares diffraction-limited images and super-resolution equivalents of the same acquisition volume to generate a quantitative map of super-resolution defects, guiding researchers in optimizing imaging parameters. The algorithm corrects for spatial offsets, estimates the resolution scaling function to convert super-resolution images into diffraction-limited equivalents, and calculates the pixel-wise absolute difference to generate an error map. We also introduce two global image quality metrics: the resolution-scaled error and the resolution-scaled Pearson coefficient. Using simulated and experimental data, we demonstrate SQUIRREL’s capacity to identify and quantify defects in super-resolution images across various imaging modalities.
Fast live-cell conventional fluorophore nanoscopy with ImageJ through super-resolution radial fluctuations, Gustafsson N, Culley S, Ashdown G, Owen DM, Pereira PM , Ricardo Henriques, Nat. Communications (2016).
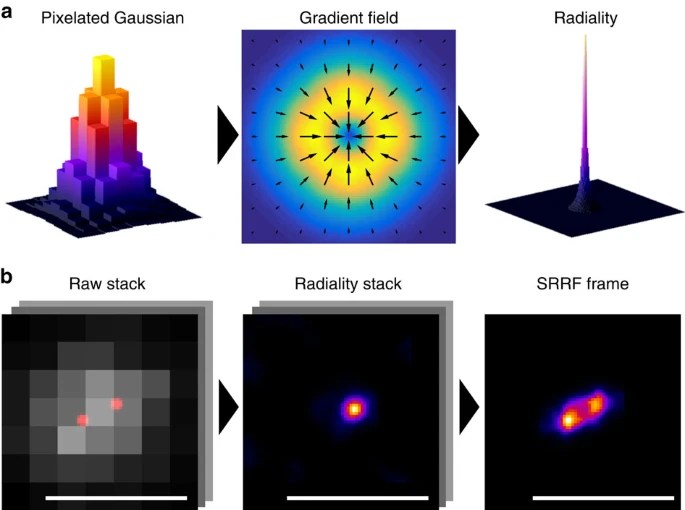
What is this about? We developed a novel analytical approach called super-resolution radial fluctuations (SRRF) that enables fast live-cell super-resolution imaging using conventional fluorophores. SRRF is provided as a GPU-enabled ImageJ plugin and achieves resolutions comparable to structured illumination microscopy (around 150 nm) using widefield, confocal, or TIRF microscopes with illumination intensities orders of magnitude lower than methods like PALM, STORM, or STED. We demonstrate SRRF’s broad applicability by super-resolution live-cell imaging of various cellular structures over timescales ranging from minutes to hours.
VirusMapper: open-source nanoscale mapping of viral architecture through super-resolution microscopy, Gray RDM, Beerli C, Pereira PM , Scherer KM, Samolej J, Bleck CKE, Mercer J, Henriques, Sci. Rep. (2016).
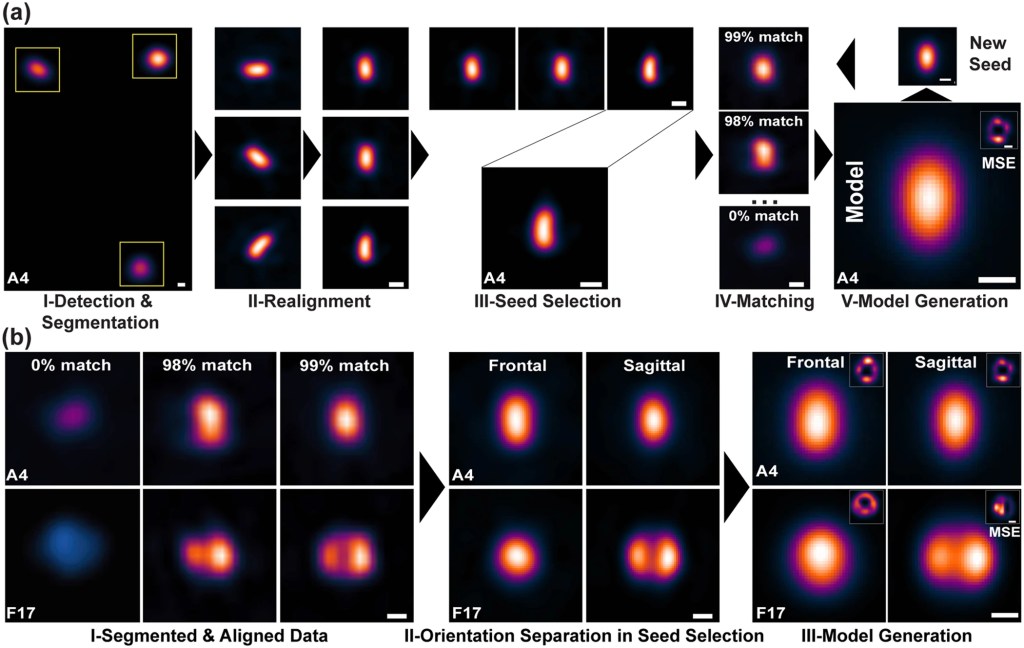
What is this about? We developed VirusMapper, an open-source software package that combines super-resolution imaging and naïve single-particle analysis to generate unbiased molecular models of viral structures. VirusMapper is a high-throughput, user-friendly, ImageJ-based tool that automatically performs statistical mapping of conserved multi-molecular structures, such as viral substructures or intact viruses. We demonstrated VirusMapper’s capabilities by applying it to SIM and STED images of vaccinia virus in isolation and when engaged with host cells. VirusMapper enables the generation of accurate, high-content, molecular-specific virion models and the detection of nanoscale changes in viral architecture.
High-content 3D multicolor super-resolution localization microscopy, Pereira PM, Almada P, Ricardo Henriques, Methods Cell Biology (2015).
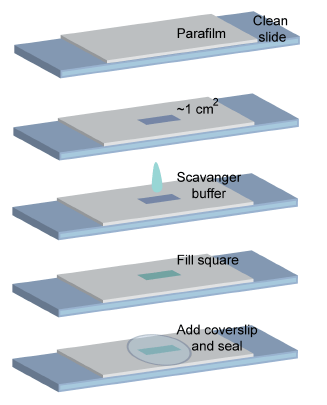
What is this about? We developed a protocol for high-content 3D multicolor super-resolution localization microscopy that combines the use of spectral demixing and point spread function (PSF) engineering to achieve 3D imaging over a large axial range. Our approach enables the reconstruction of multiple cellular structures in 3D with high sampling density using sequential labeling and imaging of targets with organic dyes. We demonstrate the potential of this technique by imaging a variety of cellular structures, including the membrane, nucleus, mitochondria, and microtubules, with high resolution in all three dimensions.
Cell shape dynamics during the staphylococcal cell cycle, Monteiro JM, Fernandes PB, Vaz F, Pereira AR, Tavares AC, Ferreira MT, Pereira PM, Veiga H, Kuru E, VanNieuwenhze MS, Brun YV, Filipe SR, Pinho MG, Nat. Communications, (2015).
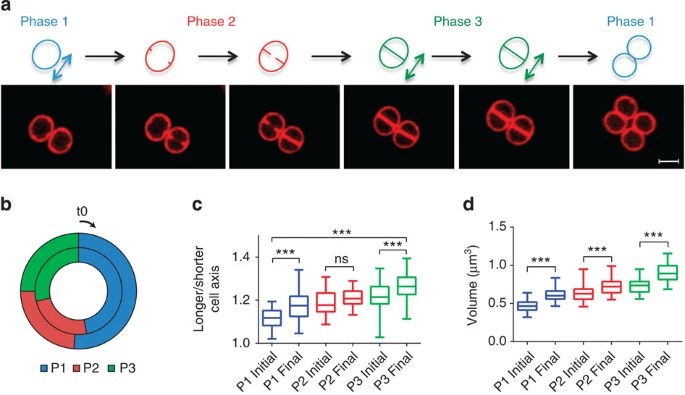
What is this about? We used super-resolution microscopy to study the dynamics of cell shape in the bacterial pathogen Staphylococcus aureus. Contrary to previous assumptions, we found that S. aureus cells are not truly spherical, but instead elongate during specific stages of the cell cycle through peptidoglycan synthesis and remodeling. We demonstrated that both peptidoglycan hydrolysis and turgor pressure are required during cell division to reshape the flat division septum into a curved surface. Interestingly, the septum generates less than one hemisphere of each daughter cell, a trait we show is common to other cocci. This results in cell surface scars from previous divisions that do not divide the cells into quadrants, introducing asymmetry in daughter cells. Our findings challenge existing models of division plane selection in cocci and highlight the need to reassess our understanding of the cell cycle in these bacteria.
Reduction of the peptidoglycan crosslinking causes a decrease in stiffness of the Staphylococcus aureus cell envelope, ±Loskill P, ±Pereira PM , Jung P, Bischoff M, Herrmann M, Pinho MG, Jacobs K, Biophysical J. (2014).

What is this about? We used atomic force microscopy to probe the effect of reducing peptidoglycan crosslinking on the stiffness of the Staphylococcus aureus cell envelope. By studying mutants lacking the non-essential transpeptidase PBP4, which is required for the high levels of peptidoglycan crosslinking in S. aureus, we found that the absence of PBP4 resulted in reduced cell wall stiffness. This effect was observed in both community-acquired and hospital-acquired MRSA strains, indicating that high levels of peptidoglycan crosslinking modulate the overall mechanical properties of the S. aureus cell envelope in clinically relevant strains.
Bacterial autolysins trim cell surface peptidoglycan to prevent detection by the Drosophila innate immune system, ±Atilano ML, ±Pereira PM , Vaz, F, Catalão MJ, Reed P, Grilo IR, Sobral RG, Ligoxygakis P, Pinho MG, Filipe SR, eLife (2014).

What is this about? We discovered that autolysins from Gram-positive pathogenic bacteria play a crucial role in trimming peptidoglycan fragments on the bacterial surface, thereby preventing detection by Drosophila peptidoglycan recognition proteins (PGRPs) and subsequent activation of the innate immune response. In the absence of autolysins, PGRPs can directly bind to exposed peptidoglycan, leading to impaired bacterial virulence. Interestingly, we found that the activity of autolysins is not limited to the producing cells but can also modify the surface of neighboring bacteria, promoting the survival of the entire population during infection.
Inhibition of WTA synthesis blocks the cooperative action of pbps and sensitizes MRSA to beta-lactams, Farha MA, Leung A, Sewell EW, D’Elia MA, Allison SE, Ejim L, Pereira PM , Pinho MG, Wright GD, Brown ED, ACS Chem Biol (2012).
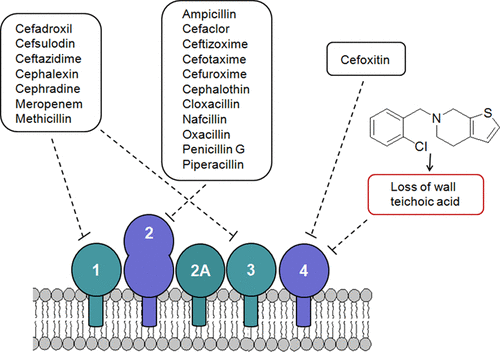
What is this about? Inhibiting the synthesis of wall teichoic acids (WTAs) in methicillin-resistant Staphylococcus aureus (MRSA) strains re-sensitizes these bacteria to various β-lactam antibiotics. By genetically deleting tarO, the first gene in the WTA biosynthetic pathway, we observed a drastic decrease in the minimum inhibitory concentrations of several β-lactams. We also found that WTA inhibition specifically blocks the cooperative action of penicillin-binding proteins (PBPs) that mediate β-lactam resistance. Our results highlight WTA synthesis as a promising target for combination therapies aimed at overcoming MRSA resistance to β-lactam antibiotics
Teichoic acids are temporal and spatial regulators of peptidoglycan crosslinking in Staphylococcus aureus, ±Atilano ML, ±Pereira PM , Yates J, Reed P, Veiga H, Pinho MG, Filipe SR, PNAS (2010).
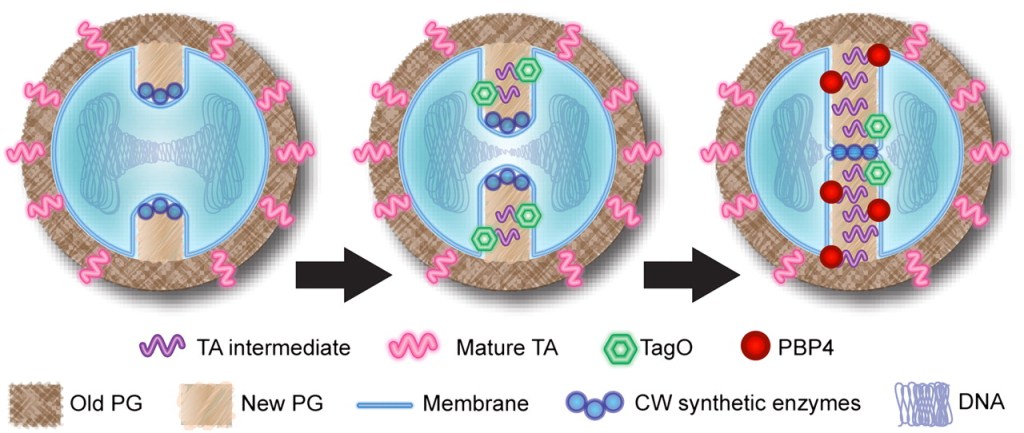
What is this about? Wall teichoic acids (WTAs) play a crucial role in regulating the temporal and spatial distribution of peptidoglycan crosslinking in Staphylococcus aureus. Using fluorescence microscopy and peptidoglycan structural analysis, we found that WTAs are required for the localization of penicillin-binding protein 4 (PBP4), a transpeptidase involved in peptidoglycan crosslinking. In the absence of WTAs, PBP4 delocalizes, leading to reduced peptidoglycan crosslinking at the septum. This delocalization is accompanied by a reduction in the percentage of highly crosslinked muropeptide species. Our findings highlight a novel role for WTAs in coordinating peptidoglycan architecture with the cell cycle, ensuring optimal crosslinking at the appropriate locations and times during cell division.

